Reini Fernandez De LucoInstitut de génétique humaine (IGH) - CNRS / Université de Montpellier
Mes recherches
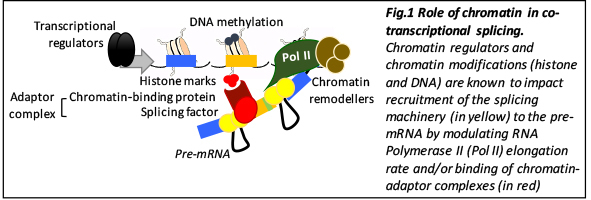
My team looks for novel mechanisms of alternative splicing regulation amongst chromatin and long non-coding RNAs. By using a well-established inducible human cell reprogramming system based on the epithelial-to-mesenchymal transition, we aim at understanding which is the dynamic interplay between chromatin, lncRNAs and alternative splicing regulation and to demonstrate its biological impact on cell-specific splicing. After a Phd in Barcelona, working on the role of chromatin on gene transcription, I moved to the NIH at Washington, DC to Tom Misteli’s lab, where I discovered the field of alternative splicing. I was amongst the first ones to demonstrate a direct role for histone marks in splicing, which was a major breakthrough for the field and allowed me to established myself in France as a young group leader with a CNRS permanent position at the IGH in Montpellier. The same year, I received the ATIP-Avenir starting grant, which was essential to set up the group and prepare my first publication as a group leader, which was the demonstration that lncRNAs can also impact splicing by creating specific chromatin signatures. This seminal work was the reason I received the CNRS bronze medal in 2016 and consolidated myself in the field. I am now expanding my research to 3D genomic interactions and its role in alternative splicing during EMT.
Mon projet ATIP-Avenir
Chromatin and miRNAs as novel regulators of alternative splicing in epithelial-to-mesenchymal transition
Alternative splicing is a highly regulated process that allows to generate protein diversity in different cell-types. Its misregulation can lead to various diseases, including cancer. Traditionally, alternative splicing has been thought to be regulated at the RNA level by combinatorial recruitment of splicing factors to specific RNA-binding sites. However, epigenetic modifications, such as DNA methylation or histone modifications, chromatin conformation, and non-coding RNAs have recently been shown to play a role in alternative splicing regulation. I hypothesize that epigenetic mechanisms may play a critical role in the establishment and maintenance of cell-specific alternative splicing programs. I propose here to characterize the function of histone modifications in alternative splicing regulation in the epithelial-to-mesenchymal transition (EMT), an important physiological process that is involved in tumor metastasis and cancer recurrence. I will first test the effect of chromatin regulators and histone modifications on the alternative splicing of two EMT-driver genes, FGFR2 and CD44. I will then extend these studies genome-wide by mapping and correlating global changes in alternative splicing with changes in histone modification levels after EMT induction. In the second part of the project, I will use a high-throughput screening approach to identify novel miRNAs and chromatin-based regulators of EMT-mediated alternative splicing. The identified candidate splicing regulators will be functionally tested for their capacity to impair EMT in human mammary cells. As a long-term goal, this project aims to uncover the regulatory role of histone marks in the definition of tissue-specific alternative splicing programs, with the ultimate goal of learning how to change splicing patterns by modulating chromatin. These studies might lead to the discovery of novel strategies to revert EMT and in this way reduce tumor metastasis and cancer progression.